── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──
✔ ggplot2 3.3.6 ✔ purrr 0.3.4
✔ tibble 3.1.7 ✔ dplyr 1.0.9
✔ tidyr 1.2.0 ✔ stringr 1.4.0
✔ readr 2.1.2 ✔ forcats 0.5.1
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
Length Class Mode
statistic 1 -none- numeric
parameter 1 -none- numeric
p.value 1 -none- numeric
conf.int 2 -none- numeric
estimate 1 -none- numeric
null.value 1 -none- numeric
stderr 1 -none- numeric
alternative 1 -none- character
method 1 -none- character
data.name 1 -none- character
Length Class Mode
statistic 1 -none- numeric
parameter 1 -none- numeric
p.value 1 -none- numeric
conf.int 2 -none- numeric
estimate 1 -none- numeric
null.value 1 -none- numeric
stderr 1 -none- numeric
alternative 1 -none- character
method 1 -none- character
data.name 1 -none- character
# one-way ANOVA # two-way ANOVA aov ( iris $ Petal.Width ~ iris $ Species , )
Call:
aov(formula = iris$Petal.Width ~ iris$Species)
Terms:
iris$Species Residuals
Sum of Squares 80.41333 6.15660
Deg. of Freedom 2 147
Residual standard error: 0.20465
Estimated effects may be unbalanced
aov ( Petal.Width ~ Species , data = iris )
Call:
aov(formula = Petal.Width ~ Species, data = iris)
Terms:
Species Residuals
Sum of Squares 80.41333 6.15660
Deg. of Freedom 2 147
Residual standard error: 0.20465
Estimated effects may be unbalanced
iris.aov <- aov ( iris $ Petal.Width ~ iris $ Species ) # Summarise the ANOVA results summary ( iris.aov )
Df Sum Sq Mean Sq F value Pr(>F)
iris$Species 2 80.41 40.21 960 <2e-16 ***
Residuals 147 6.16 0.04
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
# correlations/regressions cor.test ( cars $ speed , cars $ dist )
Pearson's product-moment correlation
data: cars$speed and cars$dist
t = 9.464, df = 48, p-value = 1.49e-12
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.6816422 0.8862036
sample estimates:
cor
0.8068949
Call:
lm(formula = speed ~ dist, data = cars)
Residuals:
Min 1Q Median 3Q Max
-7.5293 -2.1550 0.3615 2.4377 6.4179
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 8.28391 0.87438 9.474 1.44e-12 ***
dist 0.16557 0.01749 9.464 1.49e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.156 on 48 degrees of freedom
Multiple R-squared: 0.6511, Adjusted R-squared: 0.6438
F-statistic: 89.57 on 1 and 48 DF, p-value: 1.49e-12
# corresponding non-parametric tests
# A tibble: 3 × 13
# Groups: species [3]
species data test estimate estimate1 estimate2 statistic p.value
<fct> <list> <list> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Adelie <tibble> <htest> 20.4 38.8 18.3 85.8 1.87e-165
2 Gentoo <tibble> <htest> 32.5 47.5 15.0 112. 1.48e-143
3 Chinstrap <tibble> <htest> 30.4 48.8 18.4 71.1 1.18e- 75
# … with 5 more variables: parameter <dbl>, conf.low <dbl>, conf.high <dbl>,
# method <chr>, alternative <chr>
`summarise()` has grouped output by 'Treatment'. You can override using the
`.groups` argument.
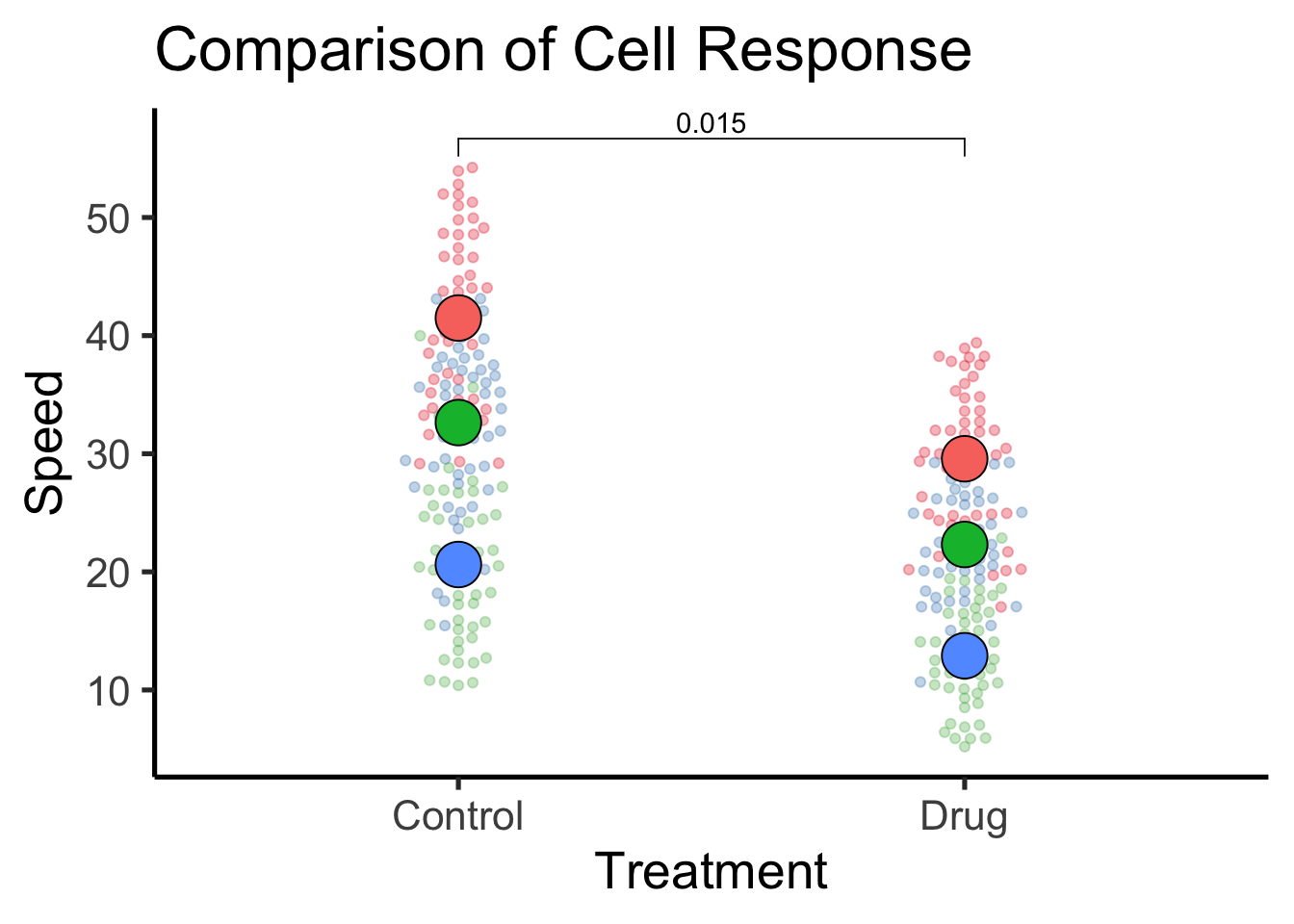
ggplot ( df , aes ( x = Treatment ,
y = Speed ,
color = factor ( Replicate )
) ) + ggbeeswarm :: geom_beeswarm ( cex = 1.5 , alpha = 0.3 ) +
scale_colour_brewer ( palette = "Set1" ) +
stat_summary (
geom = "point" ,
fun = mean ,
shape = 21 ,
mapping = aes ( fill = factor ( Replicate ) ) ,
colour = "black" ,
alpha = 1 ,
size = 8
) +
ggpubr :: stat_compare_means (
data = ReplicateAverages ,
comparisons = list ( c ( "Control" , "Drug" ) ) ,
method = "t.test" ,
paired = TRUE ,
label = "p.value"
) +
theme_classic ( base_size = 20 ) +
theme ( legend.position = "none" ) +
labs (
title = "Comparison of Cell Response"
)