import molecularnodes as mn
cv = mn.Canvas(resolution=(860, 540), transparent=True)Styles
This is how we can add styles or representations of our molecular data, so we can actually see something.
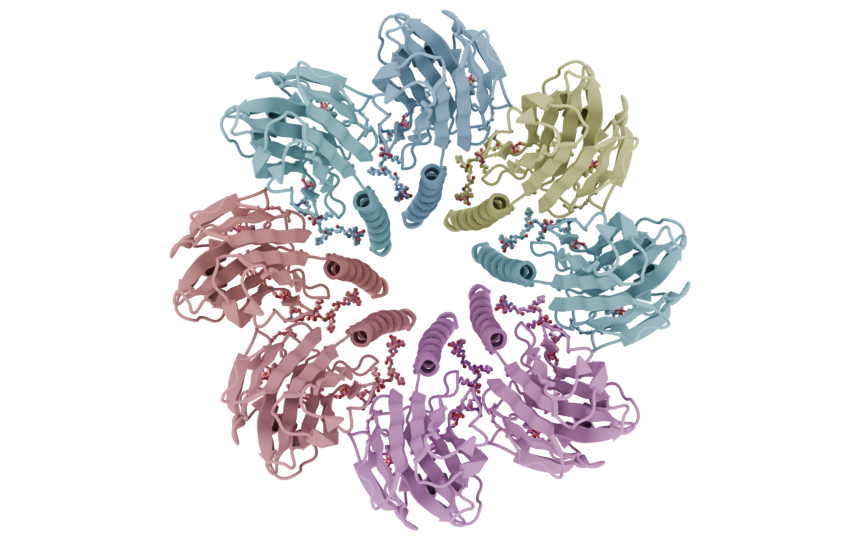
codes = ["4ozs", "8H1B", "8U8W"]
styles = [
mn.StyleCartoon(),
mn.StyleRibbon(),
mn.StyleSpheres(geometry="Instance", subdivisions=4),
]
materials = [
mn.material.Default(),
mn.material.AmbientOcclusion(),
mn.material.FlatOutline(),
]
for code, style, material in zip(codes, styles, materials):
mol = mn.Molecule.fetch(code)
mol.add_style(style, material=material)
cv.frame_object(mol)
cv.snapshot()
cv.clear()
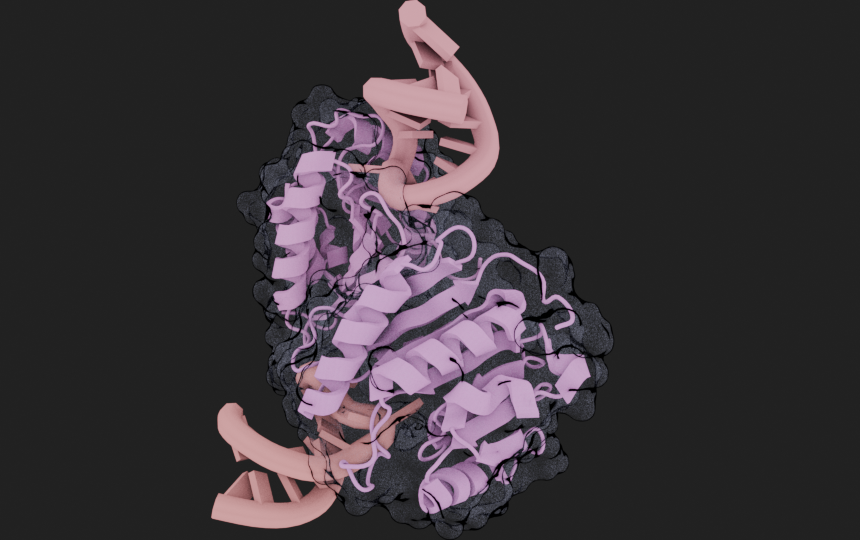

mol = (
mn.Molecule.fetch("8H1B")
.add_style("cartoon", material = mn.material.AmbientOcclusion())
.add_style(
style="surface",
selection=mn.MoleculeSelector().is_peptide(),
color=(0.6, 0.6, 0.8, 1.0),
material=mn.material.TransparentOutline()
)
)
cv.frame_object(mol)
cv.snapshot()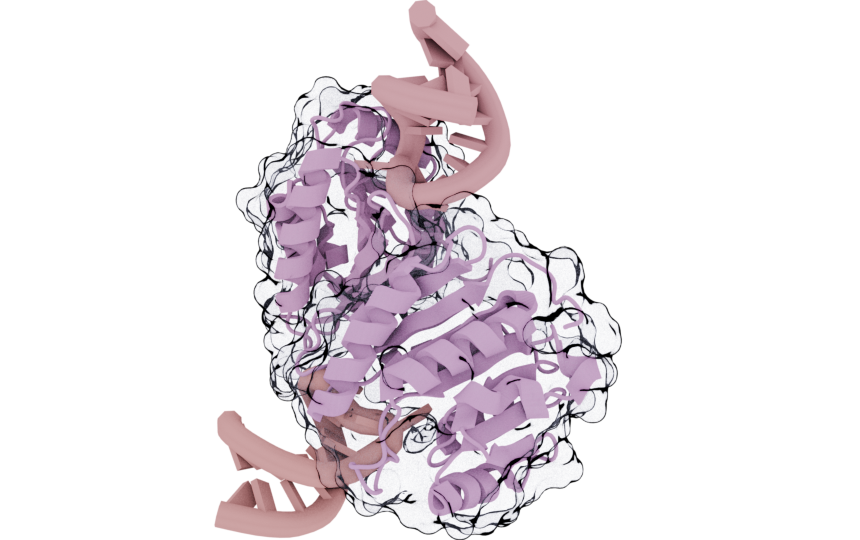
mol.styles[0].remove()
cv.snapshot()
(
mol
.add_style('ribbon', selection="is_peptide")
.add_style('surface', selection="is_nucleic")
)
style = mol.styles[1]
style.backbone_radius = 1.5
style.quality = 5
style.material = mn.material.AmbientOcclusion()
cv.snapshot()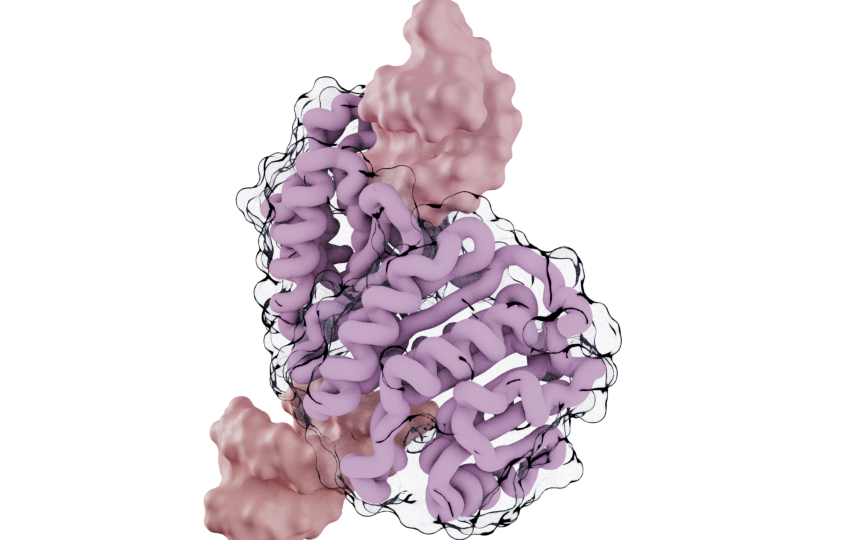
cv.clear()
cv.engine = mn.scene.Cycles(samples=32)
mol = (
mn.Molecule.fetch("9EYM")
.add_style("cartoon", material = mn.material.AmbientOcclusion(), selection = "is_peptide")
.add_style("ball_and_stick", selection=mn.entities.MoleculeSelector().not_peptide())
)
mol.styles[1].bond_find = True
cv.frame_view(mol)
cv.snapshot()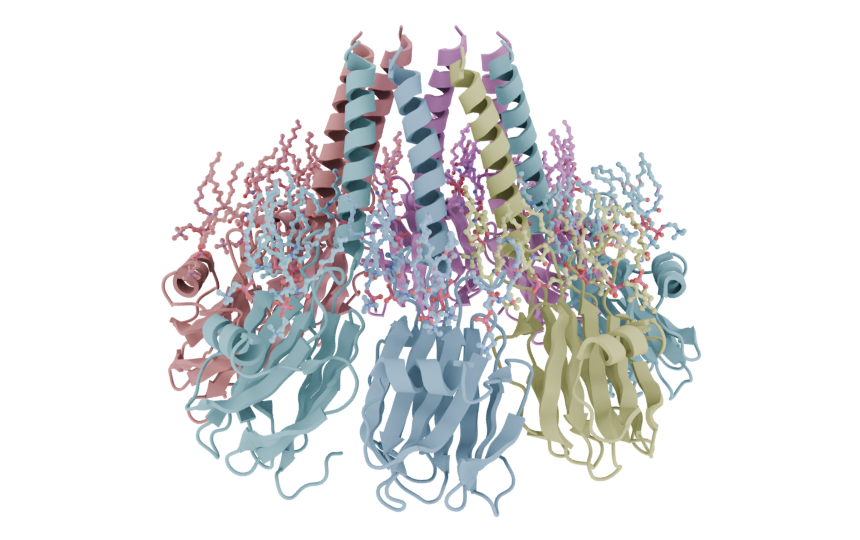
cv.frame_view(mol, (-3.14, 0, 0))
cv.snapshot()