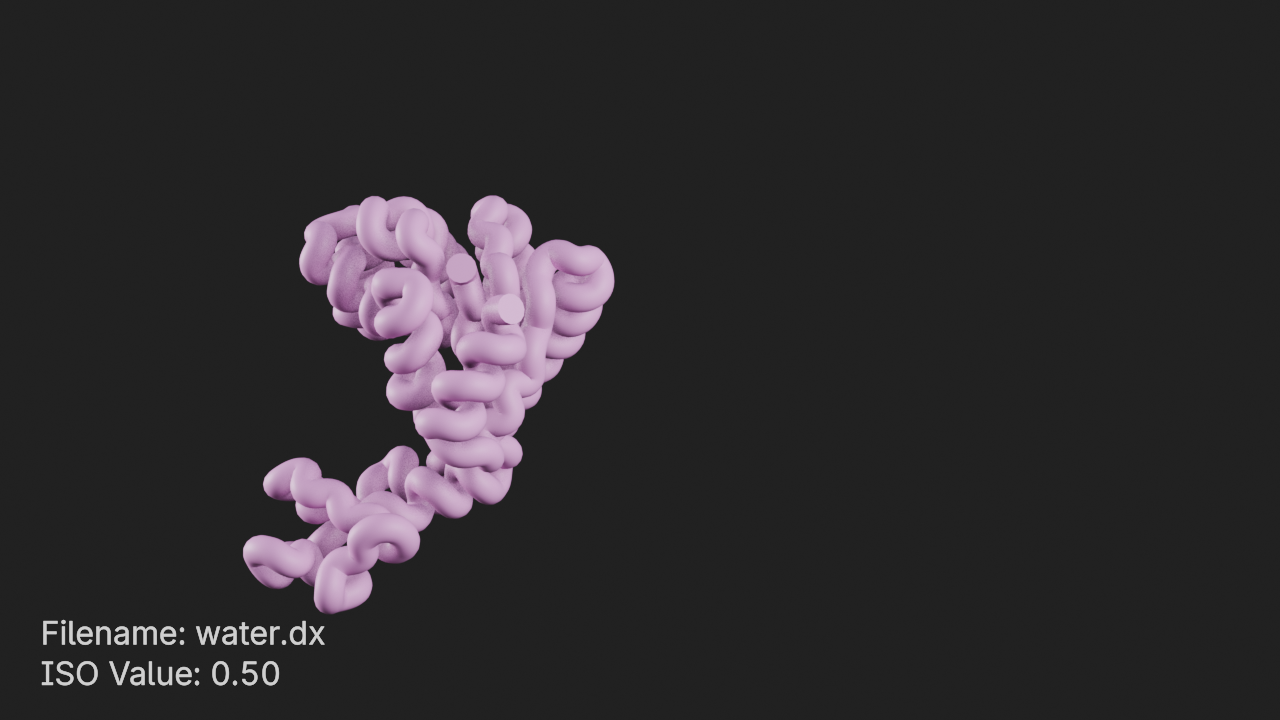
import MDAnalysis as mda
import molecularnodes as mn
import numpy as np
from MDAnalysis import transformations as trans
from MDAnalysis.analysis import density
from MDAnalysis.tests.datafiles import TPR, XTC
canvas = mn.Canvas()Solvent Density
An example for visualizing solvent density.
This example follows the Calculating the solvent density around a protein example from the MDAnalysis user guide.
Load and transform Universe
u = mda.Universe(TPR, XTC)
protein = u.select_atoms("protein")
water = u.select_atoms("not protein")
workflow = [
trans.unwrap(u.atoms), # unwrap all fragments
trans.center_in_box(
protein,
center="geometry", # move atoms to center protein
),
trans.wrap(
water,
compound="residues", # wrap each water back into box
),
trans.fit_rot_trans(
protein,
protein,
weights="mass", # align protein to first frame
),
]
u.trajectory.add_transformations(*workflow)Analyse and Export .dx
ow = u.select_atoms("name OW")
dens = density.DensityAnalysis(ow, delta=4.0, padding=2)
dens.run()
# convert density unit to TIP4P
dens.results.density.convert_density("TIP4P")
dens.results.density.export("water.dx")Add Universe to Blender
Import universe as a Trajectory and add a ribbon style to represent the protein. We also add a style to the non-rotein atoms, sliced along the y axis. After taking the snapshot for visual reference we remove the sphere style as we will be showing the water as a density.
protein_center = np.mean(protein.atoms.positions, axis=0)
t = mn.Trajectory(u).add_style(mn.StyleRibbon(quality=5, backbone_radius=2))
t.add_style(
mn.StyleSpheres("Instance", subdivisions=4),
selection=f"not protein and prop y >= {protein_center[1]}",
)
canvas.frame_view(t, (np.pi / 2, 0, np.pi / 3))
canvas.snapshot()
t.styles[-1].remove()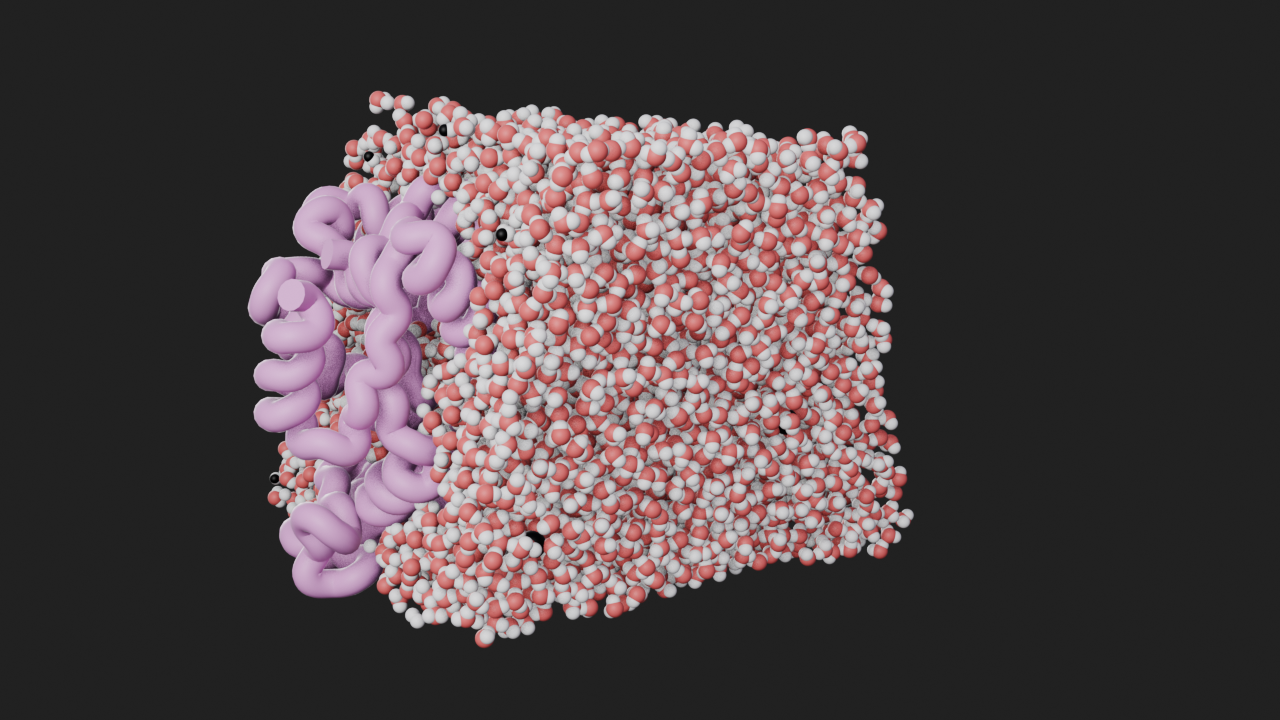
Add density component
We can load the density that was written-out from the analysis performed previously as a Density entity.
# load density file
d = mn.entities.density.io.load(
file_path="water.dx",
style="density_iso_surface",
overwrite=True,
)
# add a density info annotation for the density entity
da = d.annotations.add_density_info()
# only show the filename and ISO value
da.show_origin = da.show_delta = da.show_shape = FalseVisualization
Set density style values
# get the density style
ds = d.styles[0]
# set the positive color to blue with 50% opacity
ds.positive_color = (0, 0, 1, 0.5)ISO Value 0.5
# set ISO value to 0.5
ds.iso_value = 0.5# frame the density component and render
canvas.frame_view(d, viewpoint="front")
canvas.snapshot()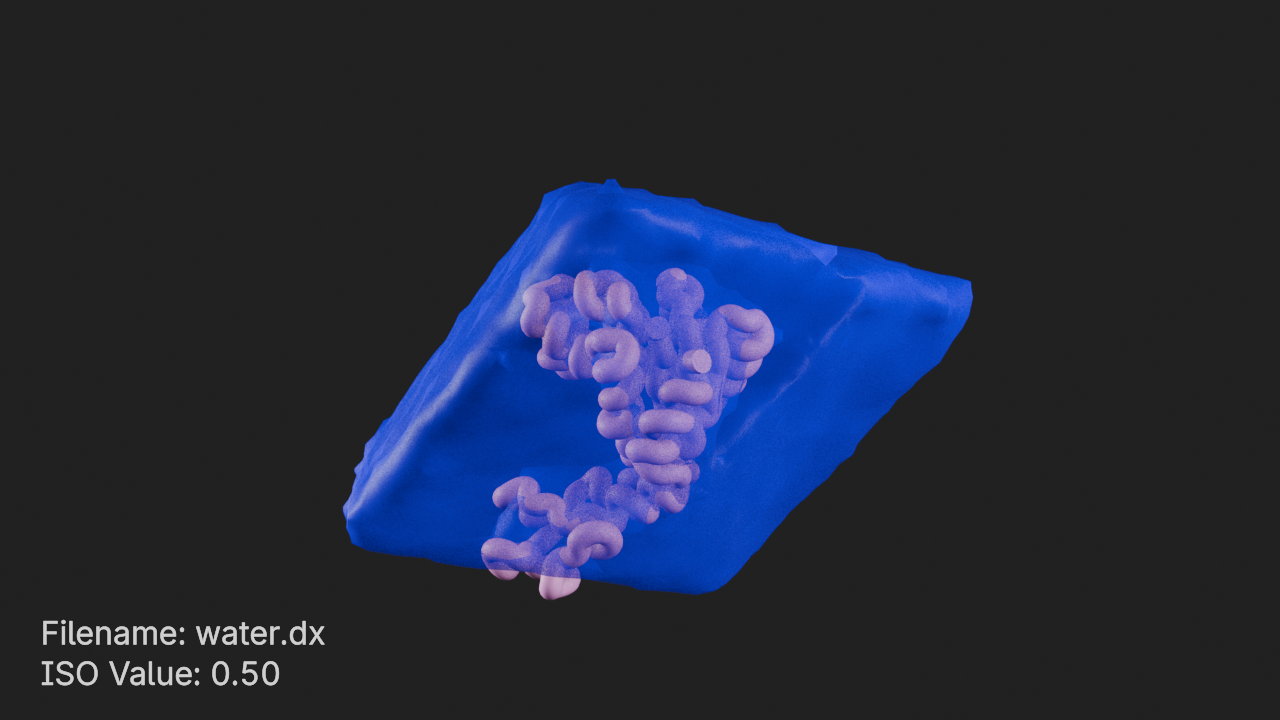
ISO Value 0.5 with Contours
# set ISO value to 0.5
ds.iso_value = 0.5
# enable contours
ds.show_contours = True
# set contour thickness
ds.contour_thickness = 0.25
# set contour colors
ds.contour_color = (1, 1, 1, 1)# frame the density component and render
canvas.frame_view(d.get_view(), viewpoint="front")
canvas.snapshot()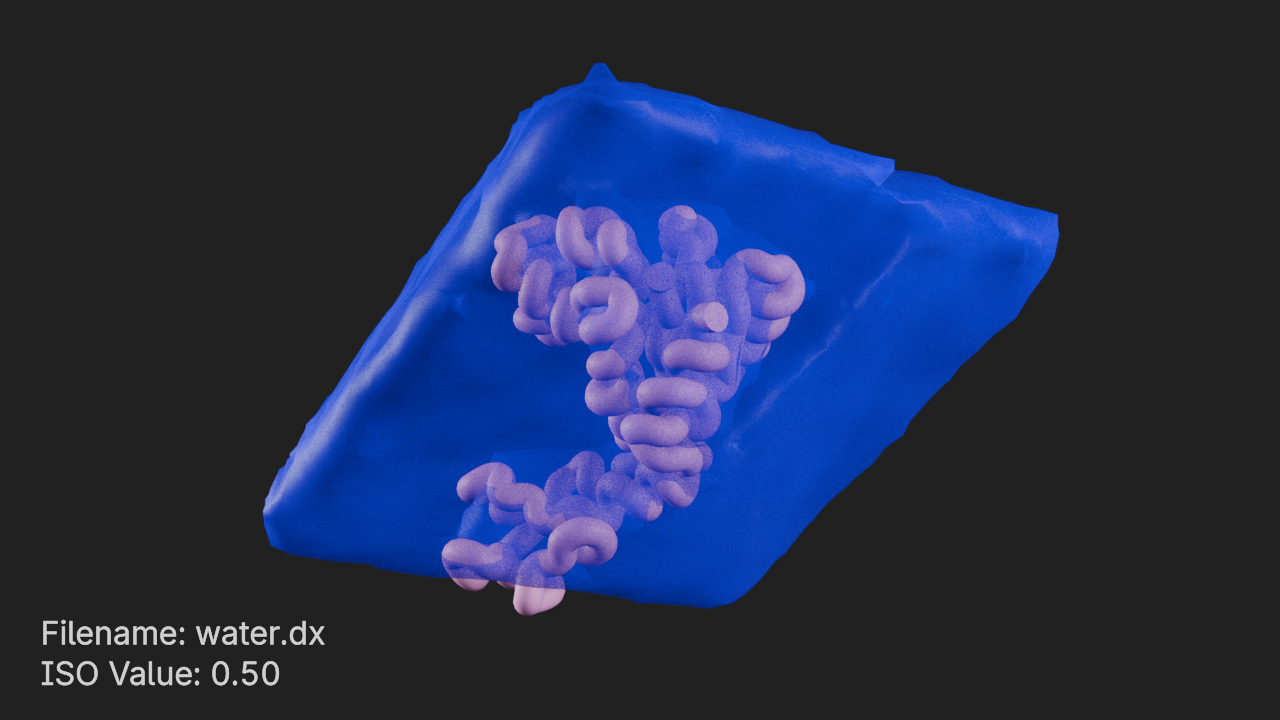
From Top with Grid Axes
# add grid axes annotation
ga = d.annotations.add_grid_axes_3d()# set viewpoint to top
canvas.frame_view(d.get_view(), viewpoint="top")
canvas.snapshot()
# hide grid axes
ga.visible = False
ISO Value 0.5 sliced from Left
# slice the grid from the left 50%
ds.slice_left = 50# set viewpoint to left
canvas.frame_view(d.get_view(), viewpoint="left")
canvas.snapshot()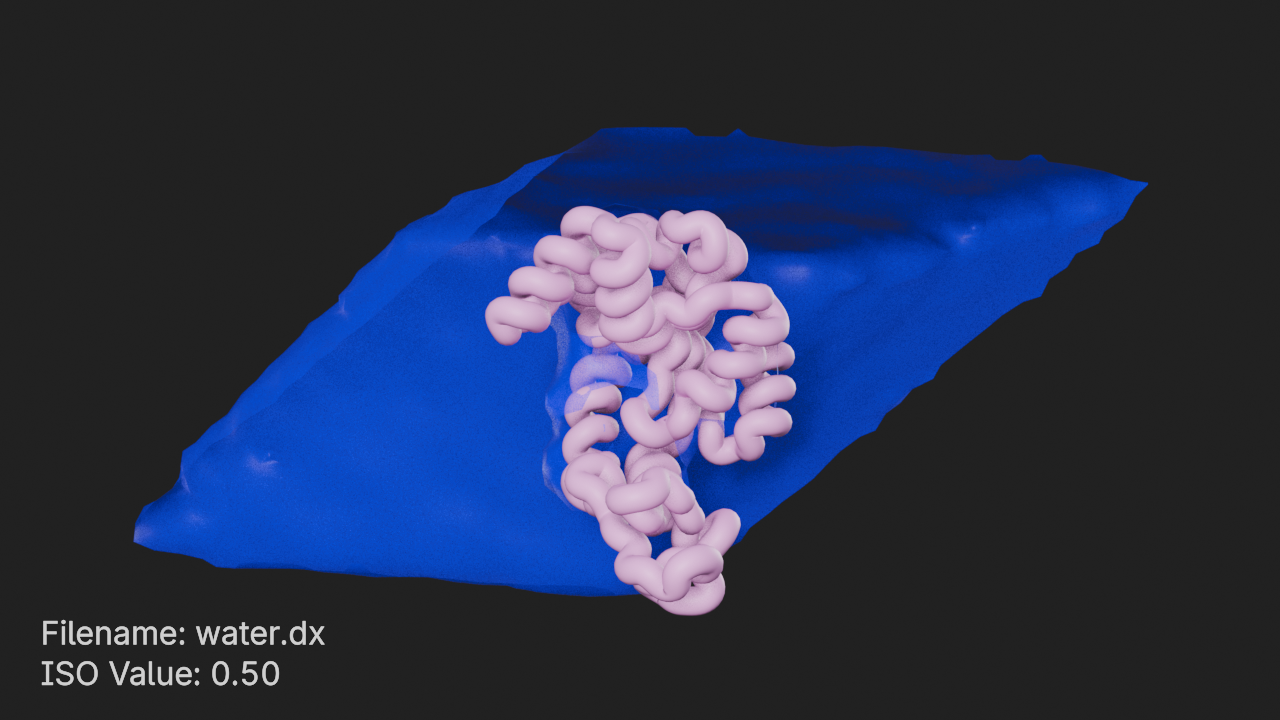
Only Contours
# reset slicing
ds.slice_left = 0
# only show contours
ds.only_contours = True# set viewpoint to front
canvas.frame_view(d.get_view(), viewpoint="front")
canvas.snapshot()