import molecularnodes as mn
import MDAnalysis as mda
from MDAnalysis.tests.datafiles import PSF, DCD
canvas = mn.Canvas()Rendering
An example of rendering MDAnalysis Trajectories.
Here is an example that uses Canvas to render images and animations of MDAnalysis Universes.
Add Universe to Blender
u = mda.Universe(PSF, DCD)
t = mn.Trajectory(u).add_style("cartoon")
# add resid 1 and 129 with spheres style
t.add_style('spheres', selection="resid 1 129")
# set the sphere geometry to mesh
t.styles[1].geometry = "mesh"Add annotations to Universe
# add universe info annotation
t.annotations.add_universe_info()
# add COM distance between resid 1 and 129 annotation
t.annotations.add_com_distance(
selection1="resid 1", selection2="resid 129", text1="r1", text2="r129"
)<molecularnodes.annotations.manager.COMDistance_interface at 0x343ec6c50>Render Image
# frame the trajectory
canvas.frame_object(t)
# render image of scene at frame 75
canvas.snapshot(frame=75)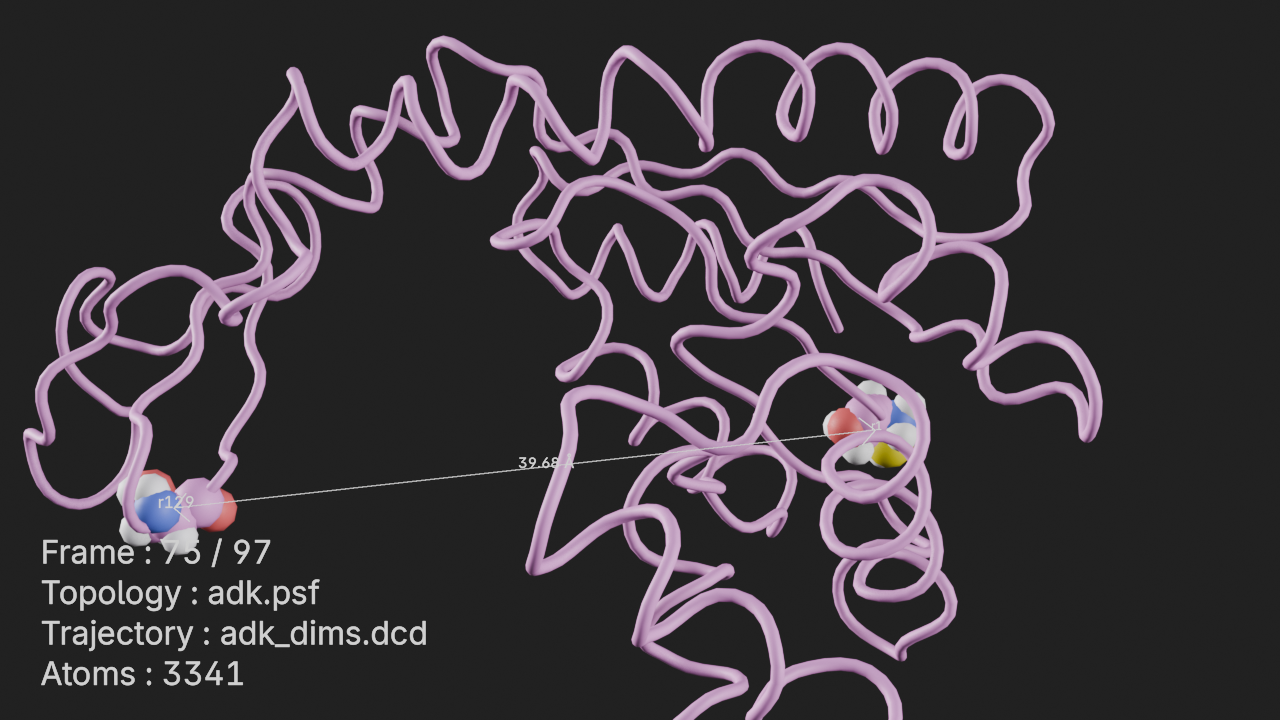
Render Animation
# render animation of scene from frame 10 to 50 at 50% sccale
canvas.animation(frame_start=10, frame_end=50, render_scale=50)